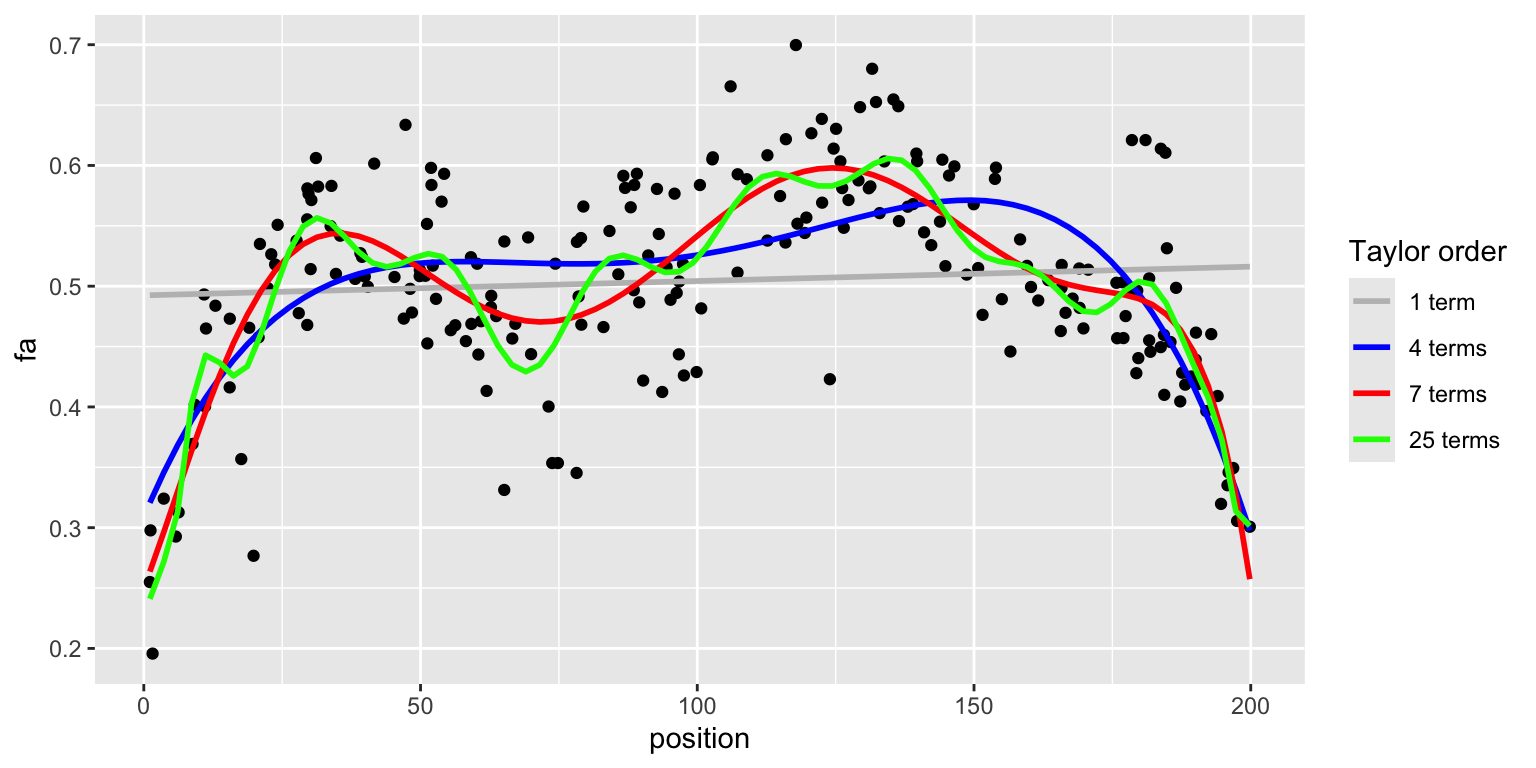
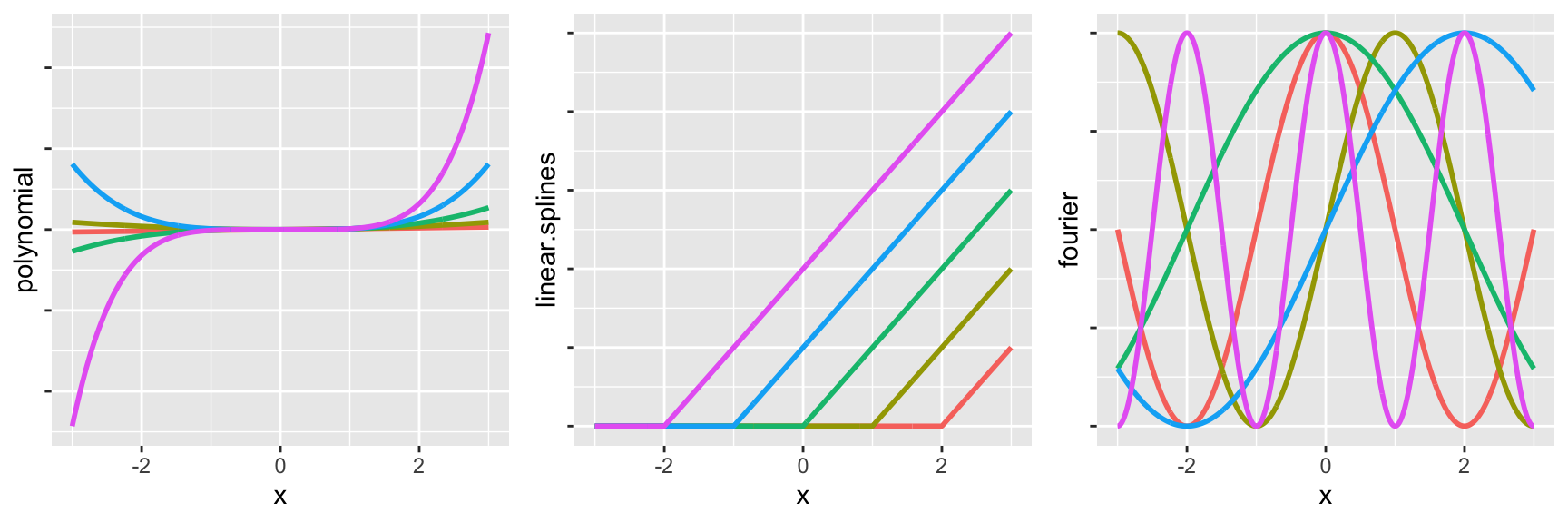
library(cowplot)
library(ggplot2)
relu_shifted <- function(x, shift) {pmax(0, x - shift)}
# Create a sequence of x values
x_vals <- seq(-3, 3, length.out = 1000)
# Create a data frame with all the shifted functions
data <- data.frame(
x = rep(x_vals, 5),
polynomial = c(x_vals, x_vals^2, x_vals^3, x_vals^4, x_vals^5),
linear.splines = c(relu_shifted(x_vals, 2), relu_shifted(x_vals, 1), relu_shifted(x_vals, 0), relu_shifted(x_vals, -1), relu_shifted(x_vals, -2)),
fourier = c(cos(pi / 2 * x_vals), sin(pi / 2 * x_vals), cos(pi / 4 * x_vals), sin(pi / 4 * x_vals), cos(pi * x_vals)),
function_label = rep(c("f1", "f2", "f3", "f4", "f5"), each = length(x_vals))
)
# Plot using ggplot2
g1 <- ggplot(data, aes(x = x, y = polynomial, color = function_label)) +
geom_line(size = 1, show.legend=FALSE) +
theme(axis.text.y=element_blank())
g2 <- ggplot(data, aes(x = x, y = linear.splines, color = function_label)) +
geom_line(size = 1, show.legend=FALSE) +
theme(axis.text.y=element_blank())
g3 <- ggplot(data, aes(x = x, y = fourier, color = function_label)) +
geom_line(size = 1, show.legend=FALSE) +
theme(axis.text.y=element_blank())
plot_grid(g1, g2, g3, ncol = 3)