dat <- read.table("data/fallacy.dat", header = TRUE, sep = ",")3 Cross-validation concerns
In this document we study how to perform cross-validation when the model was selected or determined using the training data. Consider the following synthetic data set
This is the same data used in class. In this example we know what the “true” model is, and thus we also know what the “optimal” predictor is. However, let us ignore this knowledge, and build a linear model instead. Given how many variables are available, we use forward stepwise (AIC-based) to select a good subset of them to include in our linear model:
library(MASS)
p <- ncol(dat)
null <- lm(Y ~ 1, data = dat)
full <- lm(Y ~ ., data = dat) # needed for stepwise
step.lm <- stepAIC(null, scope = list(lower = null, upper = full), trace = FALSE)Without thinking too much, we use 50 runs of 5-fold CV (ten runs) to compare the MSPE of the null model (which we know is “true”) and the one we obtained using forward stepwise:
n <- nrow(dat)
ii <- (1:n) %% 5 + 1
set.seed(17)
N <- 50
mspe.n <- mspe.st <- rep(0, N)
for (i in 1:N) {
ii <- sample(ii)
pr.n <- pr.st <- rep(0, n)
for (j in 1:5) {
tmp.st <- update(step.lm, data = dat[ii != j, ])
pr.st[ii == j] <- predict(tmp.st, newdata = dat[ii == j, ])
pr.n[ii == j] <- with(dat[ii != j, ], mean(Y))
}
mspe.st[i] <- with(dat, mean((Y - pr.st)^2))
mspe.n[i] <- with(dat, mean((Y - pr.n)^2))
}
boxplot(mspe.st, mspe.n, names = c("Stepwise", "NULL"), col = c("gray60", "hotpink"), main = "Wrong")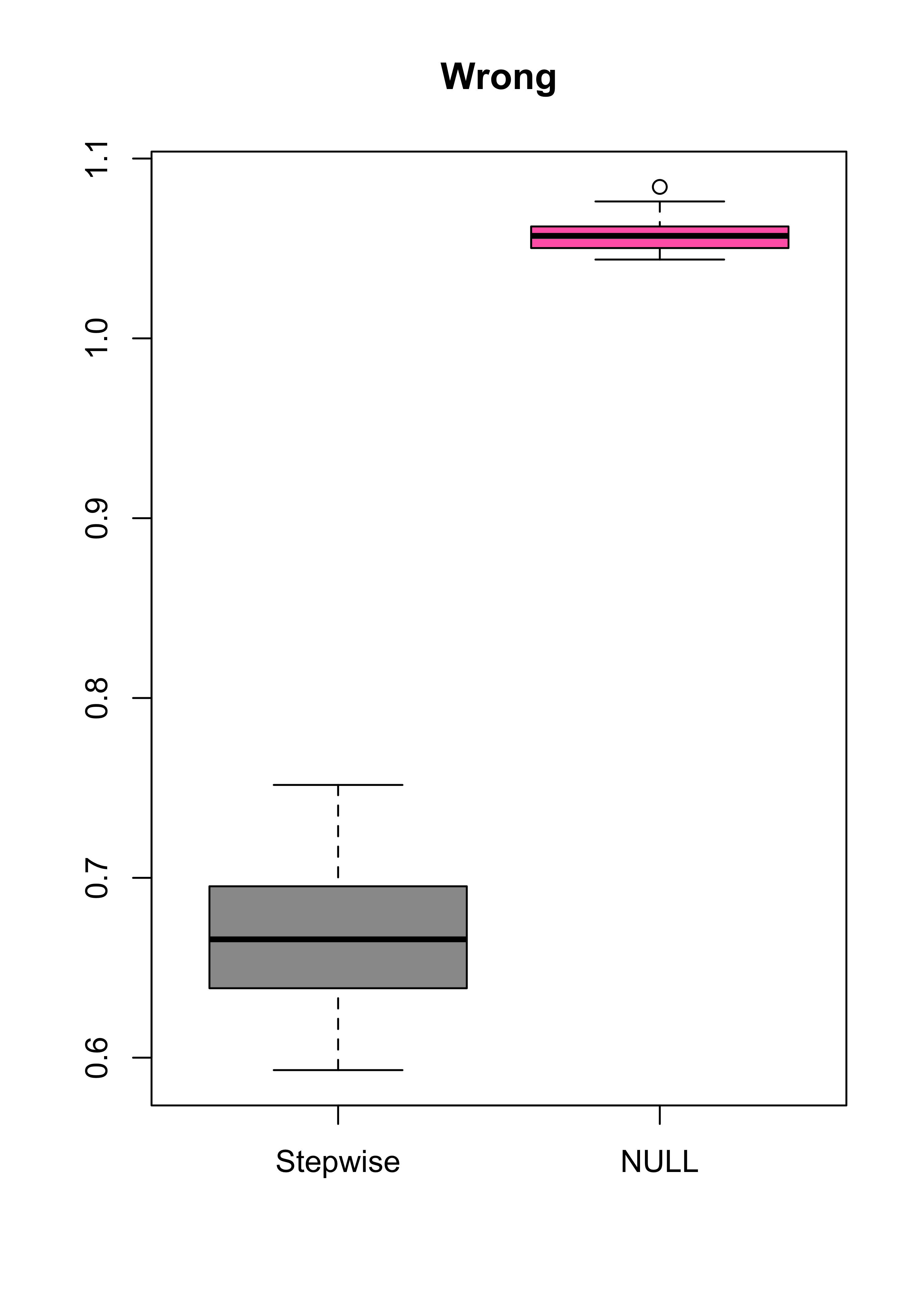
summary(mspe.st)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.5931 0.6392 0.6658 0.6663 0.6945 0.7517
summary(mspe.n)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 1.044 1.050 1.057 1.057 1.062 1.084- Something is wrong! What? Why?
- What would you change above to obtain reliable estimates for the MSPE of the model selected with the stepwise approach?
3.1 Estimating MSPE with CV when the model was built using the data
Misuse of cross-validation is, unfortunately, not unusual. For one example see (Ambroise and McLachlan 2002).
In particular, for every fold one needs to repeat everything that was done with the training set (selecting variables, looking at pairwise correlations, AIC values, etc.)